Free translation and writing by : Dr. Franyi Sarmiento, Ph.D., Inspenet, March 21, 2022.
A team of scientists from Northwestern University have discovered key structures that can drive the process that methanotrophic bacteria carry out to convert greenhouse gas into usable fuel.
Their findings could lead to the development of human-made biological catalysts that convert methane gas into methanol.
“Methane is very strongly bonded, so it’s quite remarkable that there is an enzyme that can do this,” said Amy Rosenzweig of Northwestern University, lead author of the paper. “If we don’t understand exactly how the enzyme performs this chemical reaction, we won’t be able to design and optimize it for biotech applications.”
The enzyme, called particulate methane monooxygenase (pMMO), is a particularly difficult protein to study because it is embedded in the bacteria’s cell membrane.
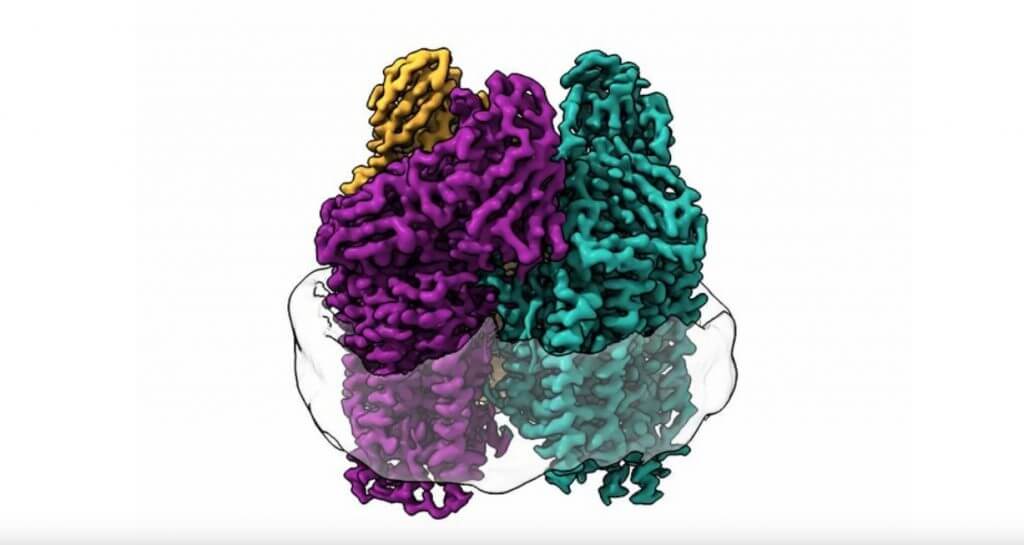
Typically, when researchers study these methanotrophic bacteria, they use a process in which proteins are stripped from cell membranes with a detergent solution. While this procedure effectively isolates the enzyme, it also removes all enzyme activity and limits the amount of information researchers can collect, such as monitoring a heart without the heartbeat.
In this study, the team used an entirely new technique. Christopher Koo used lipids from the bacteria to form a membrane inside a protective particle called a nanodisc and then embedded the enzyme in that membrane. Using cryo-electron microscopy (cryo-EM), they were able to visualize the atomic structure of the active enzyme in high resolution for the first time.
Next, the team plans to study the enzyme directly inside the bacterial cell using a cutting-edge imaging technique called cryo-electron tomography (cryo-ET). If successful, researchers will be able to see exactly how the enzyme is organized in the cell membrane, determine how it functions in its truly native environment, and learn whether other proteins around the enzyme interact with it. These discoveries would provide a key missing link for engineers.
“If you want to optimize the enzyme to plug into biofabrication pathways or consume pollutants other than methane, then we need to know what it looks like in its native environment and where the methane binds,” Rosenzweig said. “It could use bacteria with an enzyme designed to harvest methane from fracking sites or to clean up oil spills.”
The study, “Recovery of the structure and activity of particulate methane monooxygenase in a lipid bilayer,” was published in the journal Science at: https://www.science.org/doi/10.1126/science.abm3282 .
Source and internal graphic : https://news.northwestern.edu/stories/2022/03/methane-converting-bacteria/

